Synthetic biology
Synthetic biology applies engineering principles to the understanding of biological processes and the construction of cells and organisms with new function.

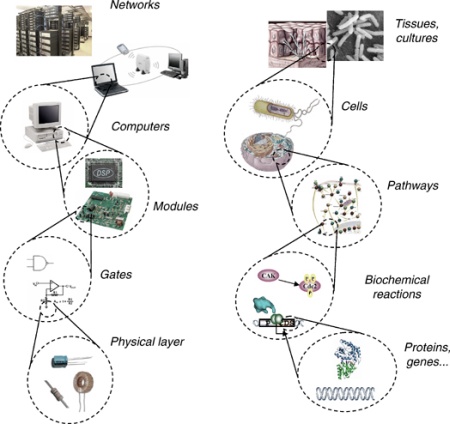
As the electronic circuitry of computers is built from transistors, processors, and mainboards, the circuitry of cells can be deconstructed into protein and gene networks. We are reprogramming cellular networks by engineering them with artificial components that allow us, for example, to conduct Boolean logic operations with DNA molecules and to expand the genetic code with unnatural amino acids. These tools enable us not only to investigate biological systems, but also to construct cells and organisms with fundamentally new functions in order to address important societal problems, especially related to human health.

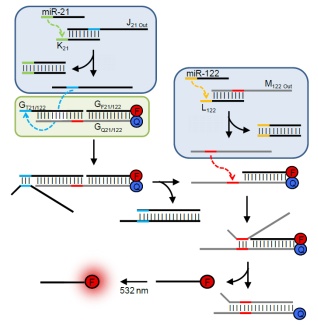
We have generated a DNA-based AND gate that responds to two microRNA input signals, miR-21 and miR-122, and entirely functions based on oligonucleotide strand displacement reactions. Only when both miRNAs are present will a quencher molecule be released from the gate, leading to fluorophore activation and miRNA detection. We have shown that these DNA logic gates are functional in human cells, opening opportunities for the development of fundamentally new, smart therapeutic agents.

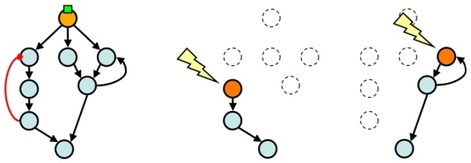
Rewiring the Cell - We rewire natural signal transduction networks used by cells to compute and respond to external stimuli. These networks perform calculations and transmit information through protein phosphorylation by kinases. We are dissecting these networks by developing optochemical biology approaches to control kinase function with spatial and temporal resolution.

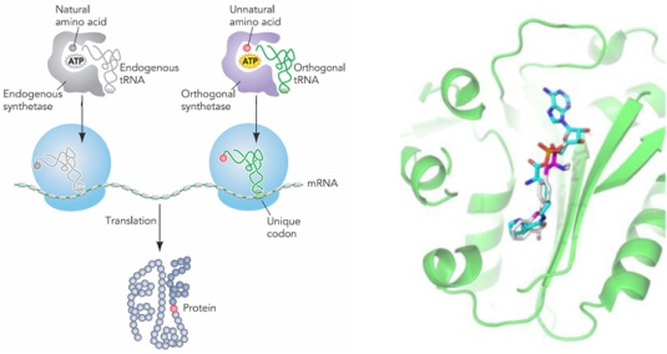
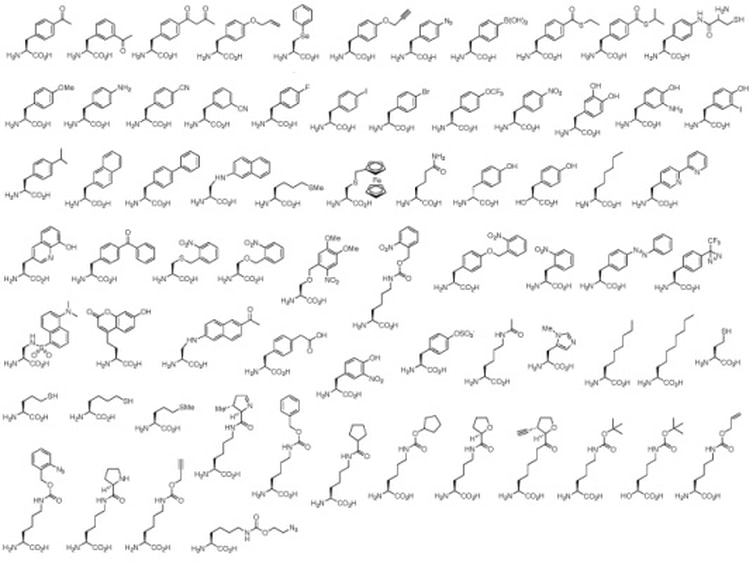
Genetic Code Expansion - We use orthogonal components, specifically an orthogonal tRNA and aminoacyl tRNA synthetase, to encode unnatural amino acids in a variety of interesting proteins. We are engineer synthetase enzymes to accept only the unnatural amino acid as a substrate for acylation of the appropriate tRNA. The unnatural amino acid is inserted into the growing protein polypeptide chain in response to a re-programmed amber stop codon.

The Scope of Unnatural Amino Acids - Hundreds of unnatural amino acids have been used to functionalize proteins. The varying structural complexity and size of these molecules often requires extensive active site engineering of the tRNA synthetases. We focus our efforts on amino acids that can be optochemically controlled, be used for bioconjugation reactions, and/or carry biophysical probes to report protein function, folding, and localization. In order to be encoded, the unnatural amino acids need to be soluble, cell permeable, non-toxic, and stable in a cellular environment.

Protein Bioconjugation - reactions often suffer from poor specificity and poor reaction rates, especially when performed in the context of live cells or organisms. We are addressing these problems through genetic code expansion with amino acids that contain reactive handles that are not found in any of the common amino acids and that allow for fast and fully bioorthogonal conjugation reactions. For example, together with the Chin lab, we genetically encoded a norbornene lysine that enabled highly selective fluorescent labeling of a cell surface protein in human cells. We are currently expanding the toolset of bioconjugation reactions to other chemistries and the labeling with other probes.